By YAN Fusheng (Staff Reporter)



With years of accumulated experience, a good hunter can tell what animals are around from the tracks or traces left by them. Now, a well-trained molecular biologist can do equally well, or even better, by interrogating a more subtle trail – the environmental DNA, or shortly eDNA, that can be from feces, mucus, skin cells, or gametes (sperms or eggs) and sampled from various environments (e.g., water, soil, the core of sediment or ice).
Over the last decade, scientists have used eDNA, particularly the genes termed “barcoding” markers within eDNA, to infer taxonomic classification and the presence of DNA-shedding animals or plants. This new technology, namely “eDNA metabarcoding,” has emerged as a new, non-invasive methodology for accessing biodiversity and illustrating ecosystems at a new level.
In principle, eDNA metabarcoding is much like what cashiers do with a barcode scanner that reads printed barcodes and gets the data shown on a screen, such as the name and price of the tagged goods.
In practice, researchers need to get to the field and collect environmental samples (e.g., water, soil, or sediment) and extract DNA from them. The purified eDNA then goes through amplification based on polymerase chain reaction (PCR) and high-throughput sequencing (HTS) to generate readouts of the barcoding markers. Finally, researchers use bioinformatic pipelines to sort readouts into groups based on sequence similarity and match them with a reference “barcoding” database to infer species presence and overall biodiversity.

A schematic workflow of environmental DNA metabarcoding that reveals species presence wherein DNA samples are collected. (Image by YAN Fusheng)
Environmental DNA for Different Fields of Study
Since the advent of commercial high-throughput sequencing platforms, scientists have started to routinely use DNA metabarcoding to access the richness of microorganisms, making it a powerful complement to conventional culture-based methods.
Then scientists extended the use of eDNA to monitor the biodiversity of macro-organisms. Given that one of its first applications was the detection of North American bullfrogs in French ponds in 2008, eDNA metabarcoding immediately drew immense attention from researchers interested in invasion biology, followed by numerous studies of eDNA sampled from various aquatic environments. These initial studies confirmed the ubiquitous presence of eDNA from aquatic animals and made eDNA metabarcoding a prevalent method in detecting invasive species and illustrating local fauna.
Environmental DNA metabarcoding enables rapid, cost-effective, and noninvasive biodiversity assessment, making it a valuable tool for conservation biology. It becomes especially handy for detecting cryptic, rare, or threatened species that pose a significant challenge for traditional physical, acoustic, or visual-based methods.
It also has the edge on the scale, speed, and comprehensiveness of informing richness and abundance of species in natural communities. eDNA can detect almost any DNA-shedding species when a comprehensive reference database is available. For example, Drummond et al. (2015) demonstrated the power of eDNA metabarcoding for accessing near-complete total richness of local fauna that covers all three domains (e.g., from bacteria to animals and plants) of life from topsoil. More importantly, scientists can use eDNA taken from the enormous scope in space and time to map the changes in species richness to inform conservation and management strategies.
Apart from informing the contemporary biodiversity, eDNA sampled from ancient environments (i.e., the core from sediment, ice, or permafrost) announces the past presence of species, because the sediment functions as a natural reservoir of eDNA. To date, animals and plants surveyed from lake sediment cores can inform about terrestrial and aquatic communities dated back to ten thousand years ago. Notably, eDNA sampled from glacial run-off may also allow us to peek into the records of animal and plant richness living in glacial and subglacial habitats.
Creative Sampling of eDNA
Scientists have also sampled eDNA creatively for many individual purposes.
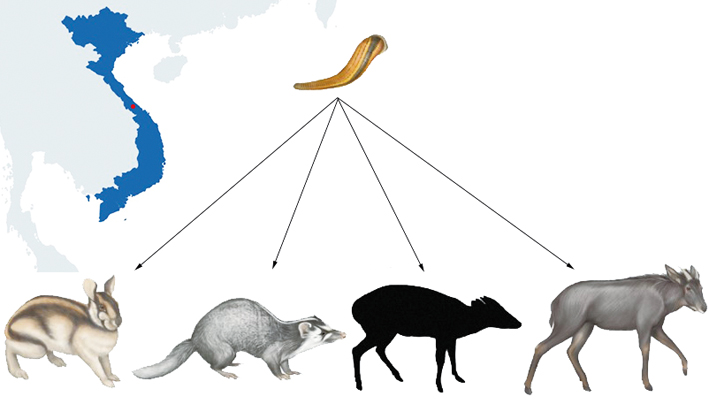
For instance, a research team from Denmark used bloodmeals from leeches to survey mammal diversity. They first demonstrated that PCR amplifiable mammalian blood DNA could survive for at least four months post-feeding in leeches and reckoned that most adult wild leeches might serve as DNA collectors of their last blood meal. The field test in a tropical Vietnamese rainforest demonstrated the method’s efficacy by testing it in situ using terrestrial leeches caught in a tropical Vietnamese rainforest and identified a cryptic, rare, and newly discovered mammalian species.

Danish researchers used bloodmeals from leeches to survey mammal diversity in a Vietnamese rainforest and identified a new cryptic species. (Adapted from I. B. Schnell et al.)
Pollen within honey or on pollinators’ bodies can be used to reveal hidden interactions between pollinators and plants. Studies have shown that the illustrations of pollination networks based on DNA metabarcoding are more complicated and informative than traditional networks based on direct observations of insect visits to plants.
Fecal DNA has also been recognized as a critical DNA source to locate predators and preys in food webs. Surveys of feces from generalist predators, such as omnivorous wild cats that cover a large hunting territory, can act as “biodiversity capsules,” and analysis of this eDNA source can indicate the biodiversity of prey communities in landscapes.
DNA plucked from air can also inform the presence of animals. Two preprints, posted on bioRxiv in July, suggest sampling air may enable a faster, cheaper way to survey nearby animals.
In December 2020, Elizabeth Clare, a molecular ecologist now at York University, set up vacuum pumps with filters in 20 locations in Hamerton Zoo Park and let them run for 30 minutes to enrich air DNA onto the filter. The team identified 25 species of mammals and birds including 17 species kept at zoo and others living near and around it, such as hedgehogs and deer.

Metabarcoding of DNA sampled from the pollen mixture carried by a honeybee can uncover its foraging preference. (Image from Pixabay)
Another team, led by Kristine Bohmann from University of Copenhagen, filtered air at three locations in Copenhagen Zoo, and detected mammal, bird, amphibian and reptile species kept at the very zoo.
It seems that the analysis of airborne DNA may offer a new way in revealing the presence of otherwise hard to detect animals, such as burrowers who live in dry environments and fleeting birds out of the sight of survey cameras.
Pros and Cons
Compared with traditional survey methods, eDNA is low-cost, noninvasive, and widely applicable for various habitats, including caves and mangroves that would otherwise be challenging for equipment or human access. eDNA also has the edge on the scale and comprehensiveness of informing species richness in natural habitats.
Additionally, one can store eDNA samples in a freezer for quite a long time and revisit them whenever needed, such as peeking on a particular family or species of organisms that draws a sudden interest. The construction of eDNA libraries over spaces and across time holds great value in conservation and management policies.

When wet mammals, including dogs and black bears, dry themselves within seconds by rapidly oscillating their bodies, they may spread DNA into the air. (Image from Pixabay)
However, like every other methodology, eDNA metabarcoding also has its limitations.
In practice, it takes some caution when interpreting eDNA data. Researchers are supposed to carefully consider the scale of inference in space and time for an eDNA sample when judging species presence in natural habitats. Such precaution stems from various uncertainties. For example, eDNA degradation can occur at different rates depending on sampling sources. Dilution is immense in oceans, quickly reducing the copy number of DNA per unit volumes to an undetectable level. Additionally, the transport of eDNA in rivers and lakes is quite different. All these factors create an inference challenge in space and time for eDNA analysis.
Based on eDNA analysis, we can not tell whether the DNA-shedding animals are alive or dead, male or female, what life stage they are in, and what they look like, which would be otherwise evident if we can hold the specimen in hands. So, as fundamentally a new way of querying the environment and sensing the biosphere, eDNA metabarcoding can supplement, not substitute, traditional biodiversity monitoring methods.
Besides, though eDNA metabarcoding has proven helpful in measuring a single species’ relative abundance in marine environments, it is still challenging to simultaneously measure the relative abundance for multiple species from eDNA. Overall, using eDNA to infer the relative abundance of different species still lacks substantial evidence. Errors can stem from the field and the laboratory, including primer bias that skews the relative abundance of amplified DNA and library preparation methods that could drive the loss of rare reads.
The Future Is Bright
Based on the above discussion, it is agreeable that eDNA has offered great opportunities in assessing community structure in different fields of study (e.g., ecology, conservation biology, and invasive biology). There are, however, many challenges of using eDNA metabarcoding as a standardized tool. The accuracy of eDNA analysis relies on continued research to improve technological aspects (e.g., amplification bias) and considerable investment in the construction of comprehensive DNA-based taxonomic reference libraries.
“Further developments associated with the impressive recent progress made in DNA sequencing technologies will allow the elimination of a DNA amplification step,” reviewed Prof. Eske Willerslev, a renowned Danish scientist who has produced numerous highly cited works on eDNA metabarcoding. In other words, it is possible to remove the errors caused by PCR amplification.
Given the power of high-throughput sequencing, building high-quality taxonomic reference libraries for eDNA metabarcoding is undoubtedly a matter of time.
As reckoned by Prof. Willerslev, “…a major challenge will be to develop new bioinformatic pipelines especially designed for exploiting such massive amounts of sequence data in the most efficient way for DNA-based species identification.”
In short, the future of using eDNA metabarcoding for ecological and biodiversity research is bright.
References
A. J. Drummond, R. D. Newcomb, T. R. Buckley, D. Xie, A. Dopheide, B. C. Potter, . . . N. Nelson, (2015) Evaluating a multigene environmental
DNA approach for biodiversity assessment. Gigascience 4, 46. doi: 10.1186/s13742-015-0086-1.
A. Pornon, C. Andalo, M. Burrus, N. Escaravage, (2017) DNA metabarcoding data unveils invisible pollination networks. Scientific Reports 7,
16828. doi: 10.1038/s41598-017-16785-5.
Christina Lynggaard, Mads Frost Bertelsen, Casper V. Jensen, Matthew S. Johnson, Tobias Guldberg Fr?slev, Morten Tange Olsen,
Kristine Bohmann, (2021) Airborne environmental DNA for terrestrial vertebrate community monitoring. bioRxiv, 2021.07.16.452634. doi:
10.1101/2021.07.16.452634
Elizabeth L. Clare, Chloe K. Economou, Frances J. Bennett, Caitlin E. Dyer, Katherine Adams, Benjamin McRobie, . . . Joanne E. Littlefair,
(2021) Measuring biodiversity from DNA in the air. bioRxiv, 2021.07.15.452392. doi: 10.1101/2021.07.15.452392.
I. B. Schnell, P. F. Thomsen, N. Wilkinson, M. Rasmussen, L. R. Jensen, E. Willerslev, . . . M. T. Gilbert, (2012) Screening mammal biodiversity
using DNA from leeches. Current Biology 22, R262. doi: 10.1016/j.cub.2012.02.058.
K. Deiner, H. M. Bik, E. M?chler, M. Seymour, A. Lacoursière-Roussel, F. Altermatt, . . . L. Bernatchez, (2017) Environmental DNA
metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology 26, 5872. doi: 10.1111/mec.14350.
P. F. Thomsen, E. Willerslev, (2015) Environmental DNA ? an emerging tool in conservation for monitoring past and present biodiversity.
Biological Conservation 183, 4. doi: 10.1016/j.biocon.2014.11.019.
P. Taberlet, E. Coissac, F. Pompanon, C. Brochmann, E. Willerslev, (2012) Towards next-generation biodiversity assessment using DNA
metabarcoding. Molecular Ecology 21, 2045. doi: 10.1111/j.1365-294X.2012.05470.x.

