By YAN Fusheng (Staff Reporter)
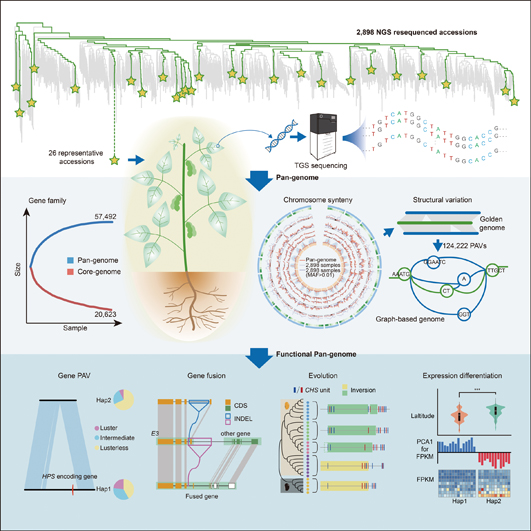
Recently, a joint team of Chinese scientists at the CAS Institute of Genetics and Developmental Biology (IGDB) constructed a new genomic map that contains almost all the genetic information of soybeans by assembling the genomes of 26 representative strains around the world into one comprehensive pan-genome. This new map is expected to benefit the upcoming efforts in soybean breeding by associating genetic variations to agronomic traits.

The comprehensive genomic map integrating the genetic information from 26 representative strains of soybean. Upper and Middle: De novo assembly of graph-based soybean pan-genome from 26 representative accessions (the items of genome sequence filed to a public database). Bottom: Functional pan-genome analyses. (Image by IGDB)
In the June 17 issue of Cell, a Chinese joint research team, led by TIAN Zhixi and LIANG Chengzhi from the CAS Institute of Genetics and Developmental Biology (IGDB), reported a high-quality graph-based soybean pan-genome. This new genomic map is expected to facilitate the gene function study in soybeans, and meanwhile help the upcoming efforts in soybean breeding.
What is Pan-genome?
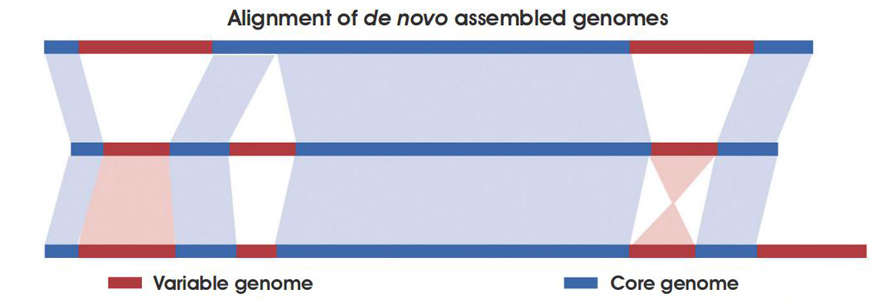
Pan-genome is a set of genomes that tends to encompass the entire gene set of all strains of a species. It entails genome sequencing of different strains, assembly and integral annotation of genomes, which give rise to a comprehensive genomic map for the species. This map tells you what genes are presented in all strains (core genome) and what genes only in some strains of a species (variable genome).
The core genome typically includes genes essential for cellular structure and fundamental metabolism. The variable genome refers to genes not present in all strains of a species, which may give an individual or a strain some signature traits that allow them to adapt to different environments. The variable genome is currently the research focus of the plant pan-genome, because it contains the clues to bridge genetic variations to biotic or abiotic resistance and many other agronomic traits.
Pan-genome is the New Fashion
Having a strain’s genome for reference opens the door to gene function study and molecular design breeding for a species; this partly explains why biologists got a long list of crops’ genomes sequenced. However, an increasing number of reports have suggested that just one or a few reference genomes cannot represent the full range of genetic diversity of a species. Therefore, pan-genome construction is becoming increasingly necessary.
As the soybean is one of the most important vegetable oil and protein feed crops, a high-quality pan-genome is required to better understand its genes account for valuable agronomic traits and harness these genes for breeding. Extensive genetic diversity among soybeans has shown the need for the construction of a complete soybean pan-genome that could cover almost all the genetic resources.
To meet this need, CAS scientists carried out in-depth resequencing and structural analyses of 2,898 soybean accessions, the filings of genomes to a public database, from major soybean-producing countries in the world, and carefully selected 26 representative soybeans, including 3 wild soybeans, 9 farm species and 14 modern cultivated varieties.
They assembled and accurately annotated the genomes of the 26 representative strains and constructed a high-quality graph-based soybean pan-genome, which contains almost all the genetic information of soybeans.
By doing pan-genome analyses, they identified numerous genetic variations that cannot be detected by direct mapping of short sequence reads onto a single reference genome. They also linked genetic variations to candidate genes that are responsible for agronomic traits (e.g., brightness and color of the seed coat), and adaptations to different environments. For example, one particular deletion in the genome associates with the soybeans’ preference for higher- or lower-latitude regions.

De novo assembly of the genomes of multiple accessions (the items of genome sequence filed to a public database) allows whole genome alignment approach to identify variable genomic regions. (Image by YAN Fusheng)
“The comprehensive information contained in this high-quality graph-based soybean pan-genome may greatly benefit the upcoming efforts in soybean breeding and research community,” highlighted the authors. This new reference might also enable scientists to reexamine the preciously acquired genomic data, and hopefully to gain insights that would not be allowed before.
Besides, increasing reports show that genomic regions outside genes, such as promotors that regulate gene expression, contribute a great deal of variance to crop traits. This suggests agronomic traits could also be determined by changes in gene regulation rather than the presence or absence of a particular genic portion. The deep understanding of how it works can provide a rich resource to mine for regulatory sequence variation that can be harnessed in breeding.
Beyond Pan-genome
“As sequencing costs continue to fall and computational power keeps rising, plant pan-genome studies are likely to expand beyond the species level such that we can start to connect pan-genomes on the genus or even the family level, allowing us to ask questions such as what gene content is required to make a legume. Eventually, this will allow us to predict and characterize the gene content of all plant species, [and to advance the] knowledge which will revolutionize future genome studies. Such wide pan-genomes will allow us to answer an age-old question: what genes make a plant?” envisioned Prof. David Edwards, a famous plant biologist from the University of Western Australia whose research focus is on the agricultural genomics, in a recent review in Nature Plants.
References
Y. Liu, H. Du, P. Li, Y. Shen, H. Peng, S. Liu, G. A. Zhou, H. Zhang, Z. Liu, M. Shi, X. Huang, Y. Li, M. Zhang, Z. Wang, B. Zhu, B. Han, C. Liang*, Z.
Tian*, (2020) Pan-genome of wild and cultivated soybeans. Cell 182, 162. doi: 10.1016/j.cell.2020.05.023.
P. E. Bayer, A. A. Golicz, A. Scheben, J. Batley, D. Edwards, (2020) Plant pan-genomes are the new reference. Nature Plants 6, 914. doi:
10.1038/s41477-020-0733-0.

