By YAN Fusheng (Staff Reporter)
Sometimes a slight difference may produce huge impact on the outcome. This is particularly true for the tiny chemical widgets added to the RNA. Recently, CAS scientists discovered that certain chemical marks on mRNA, acting as a hidden layer of gene regulation, are able to promote tumor growth and invasiveness. The findings about these chemical marks – including the nature, location and biological significance to the RNA or the cell – are valuable not only in a better understanding of the critical function of RNA modifications, but also in promising to develop new therapeutic drugs to treat human disease and even cancer.
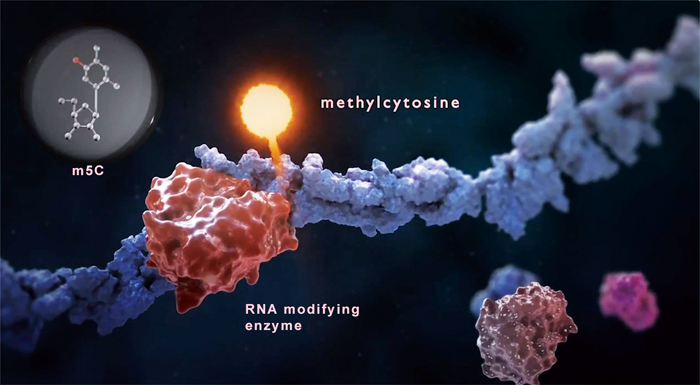
An RNA modifying enzyme adds a methyl group (lighted up in pink) to a cytosine within a strand of mRNA to give rise to a methylcytosine (m5C). Such a change may cue the cell to another fate that is different from the one when the m5C is just a normal C. (The image was adopted from the STORM Therapeutics, UK)
“When Watson and Crick discovered the double helix of DNA, it seems that we have a very simple solution to the question of how genetics works. But it turned out that it was incomplete. Because in addition to the making of the RNA, with the A, G, C and U, there were little widgets that were added to the RNA. Small chemical groups, which are not encoded by the DNA, but are added as another layer of regulation. And this is what is called epigenetics or epitranscriptomics. It’s a layer beyond just what is there in the DNA,” stated Dr. Thomas Cech, a nobel laureate and a distinguished professor from the University of Colorado Boulder.
Indeed, RNA, once thought to be a plain intermediary between DNA and protein, has now been acknowledged as a key player in regulating gene expression and cell fate. For example, the non-coding RNAs – RNA transcripts that do not encode proteins – are known to regulate many critical events during development and disease. Apart from the non-coding RNAs, chemical widgets on mRNA are now revealed to be another hidden layer of regulation that takes part in a wealth of cellular decision-makings.
RNA is not only composed of A/U/G/Cs, the four building blocks for RNA, but also of methyl-A, methyl-C and many other variations. Over 100 types of RNA epigenetic modifications have been found throughout virtually all forms of life. These RNA modifications are catalyzed by several large families of RNA-modifying enzymes. The RNA-modifying enzymes are also known as the writer proteins that “write” chemical modifications to the RNA. There are also eraser proteins that act to remove such marks. The writing or erasing of these chemical marks is often responded to a certain stimulus.
Just liking writing books, it would make no sense if no one reads them. So, it seems rational to assume that cells ought to be able to sense or read these marks, and respond accordingly. Actually, the reader proteins, or the readers, are usually the ones that carry out the biological implications of what is hold by the RNA modifications.
Two specific RNA modifications, N6-methyladenosine (m6A) and 5-methylcytidine (m5C) are now under intensive scrutiny due to their prevalent involvements in many aspects of life. Their chemical structures are as illustrated below, each bearing an additional methyl group that differs from the normal A or C. With tools to map or “read out” these specific RNA modifications, it becomes clearer that a cell needs to keep the “writing, reading and erasing” in check to function properly, and its dysregulation is often linked to disease. Up to now, scientists have confirmed that m6A plays a key role in regulating mRNA during development and disease.
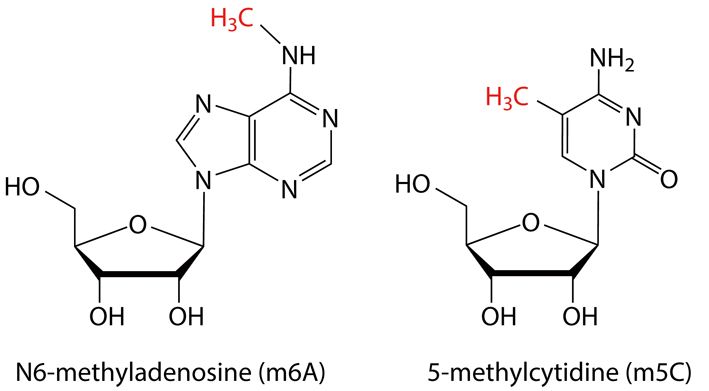
The m6A and m5C, both with an additional methyl group (in red), are currently under intensive inspection for their prevalent engagements with a wealth of biological or pathological events. (Credit: YAN Fusheng)
But how m5C takes its role under pathological conditions, especially in human solid tumor, has been largely mysterious. To address that, a joint team led by Dr. YANG Yungui from the CAS Beijing Institute of Genomics (BIG), together with collaborators from the Sun Yat-sen University Cancer Center and the CAS Center for Excellence in Molecular Cell Science/Shanghai Institute of Biochemistry and Cell Biology (SIBC) chose human urothelial carcinoma of the bladder (UCB), a certain type of bladder cancer, as disease model and revealed that tiny m5C marks on certain mRNA can control tumor cell growth and aggressiveness. This study was appeared on July 29 in the journal of Nature Cell Biology.
To Get the Big Picture
They first sought to get the big picture of how the m5C modification distributes among all RNA molecules within cells of human UCB, and whether its distribution in cancerous cells is different from that in normal bladder cells. To obtain such m5C map, they applied a certain type of RNA sequencing technique that allows to read out transcriptome-wide m5C sites at a single-base resolution. It turns out that the majority of m5C sites (around 90%) are presented in mRNA, rather than tRNA and rRNA that both are known to be replete with various RNA modifications. Despite of its origin, from either cancerous or non-cancerous cells, the m5C sites are concertedly enriched in the coding sequences and 3’ UTRs (untranslated regions) of mRNA. To better put you into perspective (see figure), a typical mRNA starts with the Cap at its 5’ end followed by the 5’ UTR, coding sequence and 3’ URT, and ends with a poly(A) tail of varied length, in which the UTRs are mainly involved with translational regulation. They also found that m5C is frequently hypermethylated in UCBs and enriched in oncogenic pathways usually linked to cancer formation. So, it seems that the distinct m5C pattern in UCBs may play a role in cancer formation and is hence worthy of a further investigation.
They next sought to find out whether the enriched m5C sites in oncogenic pathways would affect the expressions of related oncogenes and further cancer formation. To answer these questions, they scrutinized the gene expression profiles from both cancerous and normal bladder cells. As expected, they found that hypermethylated m5C sites in UCBs show different patterns of gene expression. They are either up- or down-regulated, compared with normal bladder cells. They also confirmed that several oncogenes displayed a strong correlation between m5C hypermethylation and mRNA upregulation in UCBs. The data suggested that m5C modification could enhance the mRNA level of certain oncogenes associated with UCB progression.
Bearing that in mind, scientists sought to confirm this possibility and figure out how these tiny marks make all the differences. In other words, how a cell senses the difference (m5C instead of C) and reacts in a way that promotes UCB progression.
The Reader and the Writer
As we mentioned above, a cell needs to sense the m5C marks to react accordingly. So, first thing first, they sought to pin down the reader proteins that can tell m5C from C. To hunt for such a reader, they performed an oligo pull-down assay, a strategy widely used to fish target proteins that take the “bait” (here, short strands of m5C-modified RNA oligos), and identified YBX1 as the potential m5C reader. By applying in vitro RNA-protein interaction assays, they further learned that the cold shock domain (CSD) of YBX1 preferentially recognizes m5C-modified RNA. To dissect how the YBX1 recognizes m5C, they applied structure analysis of the CSD-m5C complex and confirmed the critical role of W65, a particular amino acid in the CSD of YBX1, in delivering such m5C-reading ability.
They also found that YBX1 interacts with ELAVL1, a known mRNA-stability maintainer, and modulates the RNA-binding affinity of ELAVL1. This finding suggested a mechanism by which YBX1 recruits ELAVL1 as a “reading partner” to maintain the stability of its bound m5C-containing mRNAs.
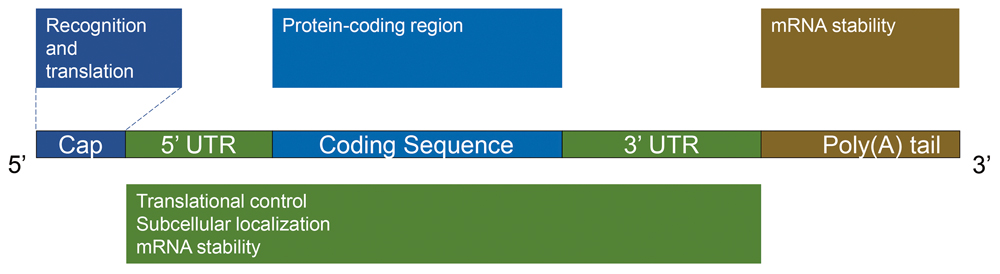
The scheme shows the structural parts for a typical mRNA, where the coding sequence is the blueprint for turning amino acids into a chain of protein (translation) and the two UTRs are mainly involved with translational control. (Credit: YAN Fusheng)
Furthermore, they found that the binding affinity of YBX1 to its target RNAs is dependent on the methyltransferase activity of NSUN2, an enzyme that gives C a methyl group to become m5C. Besides, the m5C writer NSUN2 and the m5C reader YBX1 are both aberrantly elevated in UCBs, suggesting that they could be the accomplices in promoting UCB progression. Using a patient-derived xenograft (PDX) tumor model, they demonstrated that NSUN2 or YBX1 knockdown could substantially inhibit tumor growth of UCBs in vivo. They further demonstrated that such tumor-inhibiting effect can be largely rescued by stable overexpression of wild-type NSUN2 or YBX1, but not the methyl-transferring-deficient mutant of the m5C writer NSUN2 or the m5C-binding-deficient W65A mutant of the m5C reader YBX1. Hence, they concluded that NSUN2 and YBX1 play an oncogenic role in UCB by regulating m5C. This conclusion was strongly supported by in vivo metastasis assays in mice, in which they found that NSUN2 and YBX1 promotes UCB invasiveness in an m5C-dependent manner.
How the Tiny Mark Makes All the Difference
After identifying the reader and the writer of m5C, they further sought to pin down which m5C-carrying mRNA is to be blamed and how the m5C mark on it could make all the difference to a cell.
Sieving through massive sequencing data, scientists narrowed down a possible candidate to HDGF mRNA that is a direct target of NSUN2/YBX1-mediated m5C formation or recognition. They observed an obviously shortened HDGF half-life and decreased protein levels following NSUN2/YBX1 knockdown. They also proved that reconstitution of wild-type NSUN2/YBX1, but not their mutants, could recover the HDGF protein expression in NSUN2/YBX1-depleted cells. All evidences clearly demonstrated that the sustained level of HDGF depends on both the m5C catalytic activity of NSUN2 and the m5C binding ability of YBX1.
They next determined the specific role of the HDGF m5C site in regulating its mRNA expression using a luciferase reporter assay with a gene construct containing the wild-type HDGF 3’ UTR (HDGF-WT) or 3’ UTR with a mutated m5C site (HDGF-MUT). They discovered that either NSUN2 or YBX1 knockdown substantially inhibited both the expression of luciferase mRNA and the activity of HDGF-WT but not HDGF-MUT, suggesting NSUN2 and YBX1 target the m5C of HDGF mRNA. This provides further evidence that HDGF mRNA expression requires m5C modification at its 3’ UTR specifically modulated by NSUN2 and YBX1.
Similar to NSUN2/YBX1 depletion that inhibits tumor cell growth, they also found that HDGF knockdown significantly inhibits UCB cell growth in vitro, which could be restored by overexpressing the wild-type HDGF but not the HDGF with the mutated m5C site. Assays in UCB animal models showed a similar tumor-inhibiting effect induced by HDGF knockdown as in the in vitro models. Therefore, they concluded that HDGF promotes UCB pathogenesis in an m5C-dependent mechanism.
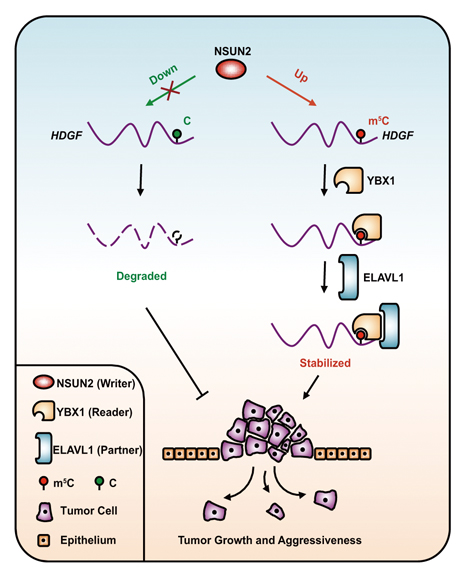
Proposed model for the regulatory landscape of the NSUN2/YBX1/m5C-HDGF signaling axis and its oncogenic role in promoting cancer pathogenesis. (Credit: Dr. YANG Yungui’s lab)
They also found that the high expression of HDGF was positively associated with advanced tumor stages and grades, and combined high expression of NSUN2, YBX1 and HDGF predicts the poorest survival in both UCB and many other types of solid tumors.
Last but not least, they also came up with a working model for the major molecular mechanism – that is, the NSUN2/YBX1/m5C-HDGF signaling axis – that promotes UCB progression. As illustrated below, the m5C hypermethylation on the oncogenic mRNA, caused by enhanced level of the writer protein NSUN2, is recognized by the reader protein YBX1 that recruits its “reading partner” ELAVL1. The binding of YBX1 and ELAVL1 stabilizes the HDGF mRNA which allows more HDGF proteins to be produced. As a result, UCB cells under this scenario display enhanced cell growth and tumor aggressiveness with feature of higher rate of metastasis. Otherwise, when this particular C is hypomethylated, no oncogene activation could occur, because the HDGF mRNA is readily degraded and unable to produce a dangerous level of the oncogenic HDGF protein.
Taken together, these results suggested that mRNA m5C hypermethylation represents a new mechanism for oncogene activation in human tumors including UCB and may serve as a valuable prognostic biomarker to distinguish UCB patients with advanced stage and/or poor survival. It also highlights the opportunity for epitranscriptomic-targeted therapy for UCB and other solid tumors.
A Race to Drug It
Growing evidences have linked RNA epigenetic modifications to cancer. The proteins that write, erase, and read these RNA modifications are now considered as promising drug targets. It has led to the launch of three biotech companies dedicated to drugging these proteins. In June 2016, Storm Therapeutics, founded by University of Cambridge scientists Tony Kouzarides and Eric Miska, raised $16 million to drug proteins that make RNA modifications. In May 2018, Accent Therapeutics, cofounded by Chuan He, Howard Chang, and Robert Copeland, raised $40 million. In October 2018, Gotham Therapeutics, cofounded by Samie Jaffrey from the Weill Cornell Medicine, launched with $54 million. A race to drug these proteins is certainly under way.
Moreover, early this year, Chuan He, a professor of chemistry at the University of Chicago who also coined the phrase “RNA epigenetics”, discovered that deleting a reader protein boosts the efficacy of cancer immunotherapy in mice. It seems that the medical relevance of RNA epigenetic modifications is ever expanding – linking m5C to cancer progression in this reported study just adds up as another demonstration. And just as Professor He says, “Epitranscriptomics is booming.”
References
Xin Chen, Ang Li, Bao-Fa Sun, Ying Yang, Ya-Nan Han, Xun Yuan, Ri-Xin Chen, Wen-Su Wei, Yanchao Liu, Chun-Chun Gao, Yu-Sheng Chen, Mengmeng Zhang, Xiao-Dan Ma, Zhuo-Wei Liu, Jun-Hang Luo, Cong Lyu, Hai-Lin Wang, Jinbiao Ma, Yong-Liang Zhao, Fang-Jian Zhou, Ying Huang*, Dan Xie*, Yun-Gui Yang*, (2019) 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nature Cell Biology 21, 978. doi: 10.1038/s41556-019-0361-y.
Ryan Cross*, (2019) Epitranscriptomics: The new RNA code and the race to drug It. Chemical and Engineering 97, 34. doi: 10.1021/cen-09707-cover.

