By SONG Jianlan (Staff Reporter)
Location matters – maybe not so much for a property as for a cell in an early mammal embryo. The position of a cell in the three-layer egg-cylinder like structure of an early mammal embryo decides what it will grow into – e.g. the heart, the gut or the brain, all of which will comprise the prospective animal body. But how does an embryonic tissue progenitor cell gradually lose its potential of developing into other types of cells? This remained largely unknown, until lately when an accurate mapping of the cell genealogy tracks the whole process in the early embryos and unravels the mechanisms underlying cell lineage determination in real time and space.
In an artist’s impression of gastrulation, which decides the blueprint that the cells will follow in their future development, the gods of fate are illustrated manipulating the fate of cells. The strings indicate the regulatory mechanisms. The illustration was a candidate for the cover image of Nature for the work described here.
(Credit: SIBCB)
The 16 cells of the mouse primordial embryo called morula know their presumable “fates” very early. During the time from the 3th to the 5th days after fertilization of the oocyte, they will gradually lose part of their developmental potential (totipotency) and be restricted to grow into certain organs (pluripotency), headed to their destination of no return. Some of them will grow into the heart, the blood and the bones, while some the lung, the liver and the intestinal tracts, and the rest the brain and the skin. By the end of the 5th day, these cells will have formed a three-layered structure called epiblast. One day later, this epiblast will commence dramatic cell movement and rearrangement, a process termed “gastrulation”. Gastrulation represents one of the prominent milestones in early embryo development, which set up the body plan for the whole animal tissues.
“It is not birth, marriage, or death, but gastrulation which is truly the most important time in your life.” Lewis Wolpert, a pioneer developmental biologist, is frequently quoted to describe how early the fate of one is determined. For cells in mice embryonic development, this process called “gastrulation” occurs only several days (5 to 9 days) after the fertilization of the oocyte. It is at this very stage that the location of a cell begins to make sense, and takes control of the cell’s fate.
Yes, the location matters a lot in the highly consolidated differentiation system. How this important process happens, however, has been elusive to scientists. How and when exactly is the “location” decided? How do the stem cells proliferate and differentiate in response to the location? Or reversely, where has a cell in a certain organ come all the way from? All these are very hard to track. Previous fate-mapping and lineage analysis had just given a vague mosaic of this location-specific allocation of “fates”. A genome-wide thorough investigation had been missing, until a joint team of biologists established a spatio-temporal molecular architecture of lineage allocation to make it possible to trace their differentiation.
The spatio-temporal progenitor lineage has resulted from an eight-year hard work originated from a brain-storming between researchers at the Shanghai Institute of Biochemistry and Cell Biology (SIBCB, also known as the Center for Excellence in Molecular Cell Science), Chinese Academy of Sciences (CAS). The work was jointly accomplished by research groups led by Prof. JING Naihe at SIBCB, Prof. HAN J. Jingdong at the CAS-MPG Partner Institute for Computational Biology, and Prof. PENG Guangdun at the Guangzhou Institutes of Biomedicine and Health (GIBH), CAS.
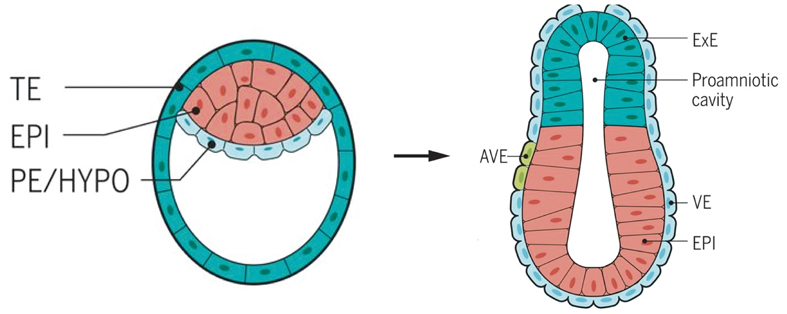
After the fertilization, the zygote cleavage divides into two, and then four, eight cells. The cells in the early embryo remain totipotent until the number of them increases to 16, and the delicate structure termed morula further expands to develop a cavity inside and becomes a blastula (left). Here the cells undergo their first differentiation some go to the embryonic side and others the more peripheral and supportive side. Later on, the one-layer blastula (left) folds inward and enlarges to become a gastrula (right), which contains three layers. Cells on the three layers each go to different destination and develop into predestined tissues and organs. TE: trophectoderm; EPI: Epiblast; PE: primitive endoderm; HYPO: hypoblast; ExE: extraembryonic ectoderm; AVE: anterior visceral endoderm; VE: visceral endoderm. (Image: Marta N. Shahbazi et al.)
“In the lab work, we sometimes found the cells developed from stem cells were not ideal, either in differentiating efficiency or in purity/functionality,” commented PENG. “This indicates our knowledge about stem cells in embryo development was inadequate. Now we have a good picture about how this process has happened in the body. The new genealogy, with both temporal and spatial information, will benefit the stem cell translational medicine and in vitro generation of organs.”
Tracing the “Allocation” at a Molecular Resolution
The highly accurate architecture depicts the spatial allocation of each cell throughout the embryonic development from the 2.5th day after fertilization, a time before the cells lose their totipotency, to the 7.5th day, when the hierarchical structure has gone deep-seated. “Here we report a spatially resolved transcriptome of cell populations at defined positions in the germ layers during development from pre- to late-gastrulation stages,” the team say in a paper published online in Nature on Aug 8 Beijing time (GMT+8).
“To define their locations to clear-enough detail at every characteristic time-points in the process, you need to profile the transcriptome of all cells in the embryo, altogether with their in situ spatial information,” said JING. Inspired by the brain storming with colleagues, he conceived of the ambitious research plan, but was frustrated by the limited precision of the technology available at that time.
Comprehensively profiling the transcriptome of a cell entails catching every transcript and accurately decoding the message they carry. RNAs are highly vulnerable to degradation in the cytoplasm – they would just disappear before one could really catch them. What made this even more demanding is, once a cell is extracted from the embryo for traditional single-cell RNA sequencing, its in situ spatial information will be lost – this method requires the sampled cells be extracted from their native tissue and randomly distributed in a single-cell suspension. Given the critical role played by the in situ locations of early embryonic cells, this would be a severe loss.
“It took Dr. PENG Guangdun nearly two and a half years to develop a pipeline for accurate spatial transcriptomic analysis,” said JING, looking at PENG, his former PhD student and now a principal investigator at GIBH. PENG has travelled all the thick and thin of this long expedition.
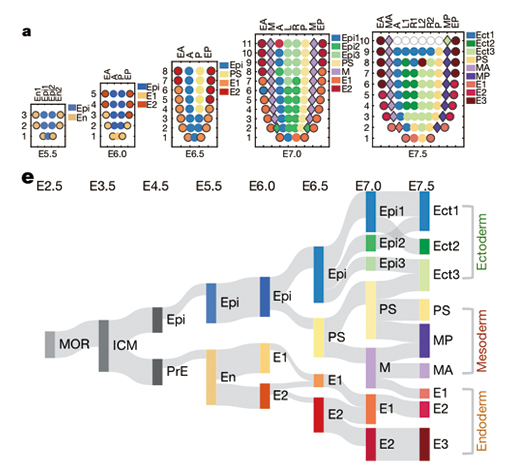
The team found the part of the cells on the endoderm could have directly come from the primitive endoderm (PrE), rather than as originally believed to have been mediated by an epiblast (Epi) stage. Forming a contrast, some cells on different “floors”, thought to be irrelevant with each other, are very likely offspring from the same precursor cells. A). The spatio-temporal locations of cells from the 5.5th day to the 7.5th day after fertilization; B). The phylogenetic tree based on the newly established molecular lineage. The thickness of the connecting bands indicates the strength of correlation between lineages.
Legend: MOR: Morula; ICM: Inner Cell Mass; Epi: Epiblast; PrE: Primitive Endoderm; En: Endoderm; PS: Primitive Streak; M: Mesoderm; Ect: Ectoderm; MP: Posterior Mesoderm; MA: Anterior Mesoderm.
(Credit: SIBCB)
PENG and his colleagues developed a method that combines laser capture microdissection (LCM) and single-cell RNA sequencing technology, to simultaneously obtain the location and transcriptome of a cell. Building on this newly developed pipeline named “Geo-seq”, the team captured cells from different spatial locations at several consecutive developmental moments, and performed transcriptome analysis – at a high sampling rate, covering the whole transcriptome. Only after all this arduous and persistent work, did they establish a precise “atlas” for the cells’ growing trajectories.
The laser of the LCM equipment knows all their way here – The analysis requires such enormous numbers of accurate sampling that the laser beam got burnt out and had to be replaced.
Now the genealogy has been accomplished. “This marks the most complete and exhaustive interactive database for spatio-temporal transcriptome of mice gastrulation in the world,” commented Profs. TANG Fuchou and ZHOU Fan at the Biomedical Pioneering Innovation Center of Peking University, China.
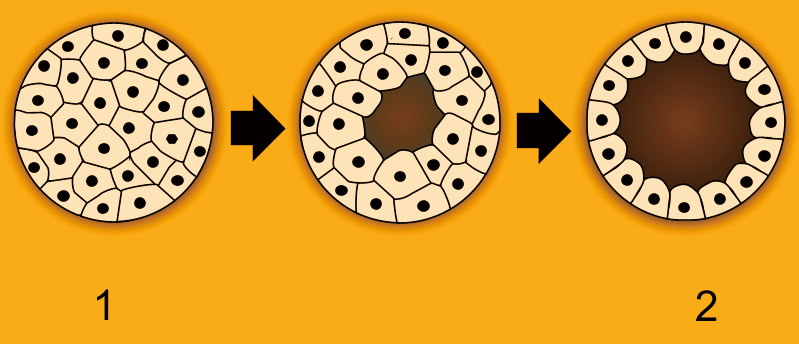
The morula (left), in which cells are totipotent, expands and gives rise to a cavity inside, and later develops into a blastula (right). At this stage the cells undergo their first differentiation: they are divided into two types, some stay outside and harden to form a supportive structure called trophoblast, and others, which are a mass of germ cells, remain inside the cavity and go onto a more “embryonic” way of no return. These pluripotent cells, called inner cell mass (ICM), will become the future three-layer gastrula, containing ectoderm, mesoderm and endoderm. (Image by Pidalka44, Wikimedia Commons, https://commons.wikimedia.org/w/index.php?curid=123497)
The team did not stop at this. For them, this refined “chart” has just paved the way for further explorations. Navigating with this long-anticipated map, they know better where to go, and have already made some discoveries, no lack of subversive ones.
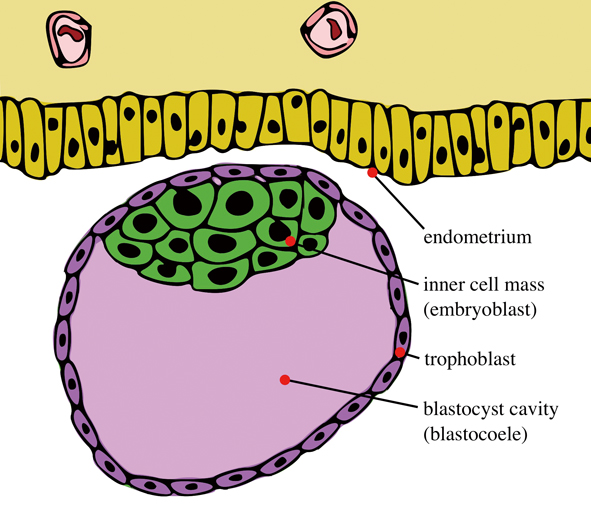
Illustrated is a blastocyst just before implantation, with an inner cell mass and trophoblast. The inner cell mass inside will become the future endoderm, mesoderm and ectoderm, according to traditional developmental theories. (Image: Seans Potato Business, CC BY-SA 3.0)
Subversive Findings
Now, tracing along this cell genealogy, the team can tell the origin of any a cell on any “room” of a complex building. They found some “dislocation” and “convergence” of cell lineages that might subvert the well-believed phylogeny of tissue lineages. “We might need to revise our textbooks,” commented Prof. ZHOU Jinqiu, vice director of SIBCB and expert in epigenetics.
It had been well established that during the gastrulation, the inner cell mass (ICM) of the blastula (derived from the morula, MOR) first expands and gives rise to the epiblast (Epi), then the latter further grows and develops into three layers of inner germ cells: the ectoderm, the mesoderm and the endoderm. In other words, all the three layers are derived from the epiblast. However, JING’s team found that very likely part of the endoderm cells could have “skipped” the epiblast stage and directly developed from the primitive endoderm (PrE). On the other hand, some cells on the mesoderm and the ectoderm, originally believed to have been from different ancestors, turned out to be offspring of a common precursor.
With unprecedented detail, the spatio-temporal transcriptome established a continuous expression profile where the researchers can follow the cell signaling network and trace back to certain genes. This offers new opportunities to explore and reconstruct mechanisms underlying the specification and tissue formation. Examining how the cell signaling pathways are activated or inhibited between different regulative factors in the transcriptome, the team discovered that Hippo-Yap signaling, known to have a role in detaching the trophectoderm from the inner cell mass during the transition from the morula to the epiblast stage, became enriched exclusively in the endoderm, suggesting its possible regulative role in early-endoderm development.
Following the signaling network underlying pluripotency, the team also established a timeline for cellular differentiation, underpinning when exactly the embryonic stem cells transit from totipotent (before their implantation into the uterine wall) to pluripotent (after transplantation), and then lose their pluripotency (late gastrulation).
Answering Fundamental Questions
“The lineage allocation answers the very ultimate three questions – who are you, where are you from, and where are you going,” commented Prof. HUI Lijian, leading developmental biologist in China and also JING’s colleague at SIBCB. “These questions are very crucial for human beings, and as well as for the stem cells. Acquiring this detailed genealogy helps us explore many previously impossible issues. For example, understanding how stem cells differentiate and how this process is regulated, makes it possible to manipulate the differentiation and artificially build organs. Knowing the genealogy of stem cells, we will also know new pathways or sources to obtain different types of them, and better understand some diseases at a molecular level,” he explained. “To a large extent, the position defines the ‘identity’ of a stem cell. Its spatial and temporal information is closely integrated and inseparable. This combination helps us predict how the cells build up an organ, or how they get the epigenetic imprints,” he continued. HUI, who took part in the brain storming that gave birth to the original idea of mapping the early embryo differentiation, felt happy to have seen the idea to grow up.
The establishment of the 4D high-resolution genealogy of lineages would greatly improve the understanding of early embryonic cell differentiation, as well as the regulation of pluripotent stem cells, experts say. This is important for research in early embryonic development as well as stem cell regenerative medicine. For example, given the key role played by cell signaling in early development, manipulation of it at this stage can induce formation of targeted tissues and hence inspire treatments or even cures for some diseases or injury cases. Rising from this newly established horizon, could be unexpected discoveries.
References
Guangdun Peng*, Shengbao Suo, Guizhong Cui, Fang Yu, Ran Wang, Jun Chen, Shirui Chen, Zhiwen Liu, Guoyu Chen, Yun Qian, Patrick P.L.Tam, Jingdong J. Han*, Naihe Jing*. (2019) Molecular architecture of lineage allocation and tissue organization in early mouse embryo. Nature 572,528–532.
Guangdun Peng, Shengbao Suo, Jun Chen, Weiyang Chen, Chang Liu, Fang Yu, Ran Wang, Shirui Chen, Na Sun, Guizhong Cui, Lu Song, Patrick P. L. Tam, Jing-Dong Jackie Han*, Naihe Jing*. (2016) Spatial Transcriptome for the Molecular Annotation of Lineage Fates and Cell Identity in Mid-gastrula Mouse Embryo.Developmental cell 36, 681–697.
Jun Chen, Shengbao Suo, Patrick. P.L. Tam, Jing-Dong J. Han, Guangdun Peng*, Naihe Jing*. (2017) Spatial transcriptomic analysis of cryosectioned tissue samples with Geo-seq. Nature protocols 12(3), 566–580.

