Scientists’ impression of circular RNAs: Ring-shape RNAs swimming in cytoplasm look just like this balmy plant of Lysimachia christinae Hance, a Chinese medical herb that helps alleviate irritations caused by infections. Due to the special double-strand secondary structures on their rings, this obscure variant of RNAs might offer new hope for curing systemic lupus erythematosus (SLE), as revealed by teams of CAS scientists in Shanghai. (Image courtesy of CHEN’s lab)
“It helps to answer a very important question in the causal pathway of lupus and offers new hope for therapeutic strategy of this common and severe autoimmune diseases,” commented Dr. SHEN Nan on a recent discovery by teams of biologists based in Shanghai.
SHEN is currently leading an institute dedicated to research and treatment of autoimmune diseases, one of the three major threats to human health named by WHO. His team has long sought in vain to discover the cause of the unprovoked overreaction of the immune system in patients with systemic lupus erythematosus (SLE), the deadliest and most frequently studied of autoimmune diseases. In the case of SLE, the immune system attacks the normal cells of the patient and causes damage to their otherwise healthy tissues and organs. No cure is available, and interference in the immune regulation could be tricky, as any mistake could harm its activity and responsiveness to future infections. Now the newly revealed role of ring-shape RNAs might help to further the pathological research into this disease and provide better understanding of its causal pathway.
This discovery was made by teams led by Dr. CHEN Lingling at the Shanghai Institute of Biochemistry and Cell Biology (SIBCB, also known as the Center for Excellence in Molecular Cell Science) and by Dr. YANG Li at the CAS-MPG Partner Institute for Computational Biology (PICB) under the Chinese Academy of Sciences (CAS). They reported in a recent issue of Cell that circular RNAs (circRNAs) could anchor the immune system via binding to a natural immune factor called PKR and curb its activity. This does not hamper the normal functioning of the immune system but keeps it quiescent; once a viral infection does occur, a biocatalyst called RNase L is activated to promote the degrading of these inhibitor RNAs and free the bound warrior PKR to fight the invaders.
Exploring possible immune implications of this suppression, researchers wondered if malfunctioning of this biological lock can induce overreaction of the immune system – the major cause of autoimmune diseases. Their ensuing cooperative research with Dr. SHEN at the Shanghai Institute of Rheumatology under the Renji Hospital (SIRRH) confirmed their speculation and helped answer an important question long-perplexing pathologists.
Once again, circRNAs emerged from obscurity and caught the attention of the scientists.
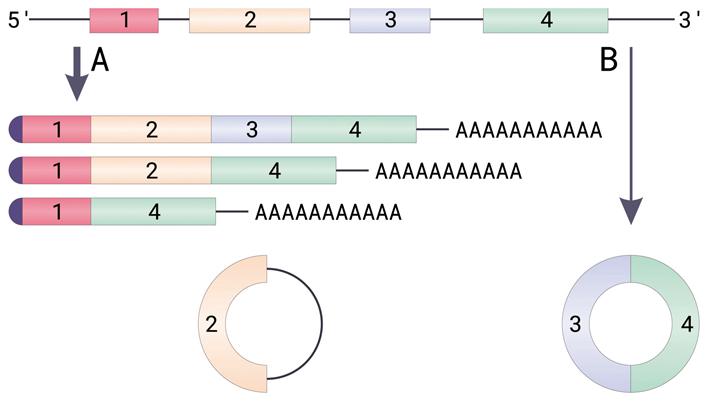
Illustrated here is the biogenesis of circular RNAs from pre-mRNAs via back-splicing events:
A. Canonical splicing of an mRNA. Due to alternative splicing, the chain can give birth to different splice variants. All these mature mRNA variants, however, have a cap at the 5’ end (presented in dark violet half-circle) and a poly-A tail (adenine nucleotides) at the 3’ end.
B. Back splicing and circRNA formation. Circular RNAs can have one or more trunks of coding sequences (exons), sometimes retaining a non-coding intron (depicted as a dark line near the 2 exon).
[Image adapted from the original work by Helixitta (CC BY-SA 4.0 https://creativecommons.org/licenses/by-sa/4.0)]
Hero the Obscure
Generally carrying no effective genetic information that codes the assembly of proteins, non-coding RNAs (ncRNAs) are not capable of producing functional proteins. Dwarfed by their “coding” counterparts, they had been long neglected in biological research until quite recently. As an unlucky part of this obscure family of negligible genome-wide detection, circRNAs only first caught the attention of scholars as a “mistake.”
They emerge at a very low rate in a process called “splicing” of cellular machinery, in which the immediate “casts” from genes, called precursor messenger RNAs (pre-mRNAs), get “refined” and ripen into mature messenger RNAs (mRNAs). The latter are to carry the message from the gene out of the nucleus, and serve as templates for protein assembly in the cytoplasm. During the process of a canonical splicing in a eukaryotic nucleus, the “redundant” fragments on the pre-mRNA chain, which do not code protein assembly, shed off and the “shortlisted” parts – those are supposed to guide the genesis of proteins – join together. Under some unusual conditions, however, the chain could fuse itself nose to end by back-splicing to form a covalently closed continuous loop and hence produces circular RNAs.
Following the codes on these heretic RNAs, the cellular machinery generally cannot produce any functioning proteins, though several exceptions came to light over the past few years. Given that their linear cousins derived from the same pre-mRNAs can be translated into proteins in the usual way, this infertile lineage acquired a reputation of mistaken products from improper splicing.
Results from the latest research are subverting this impression, thankfully. This is not because their exceptional buddies that can bear proteins just like their linear cognates, however, but because of accumulating clues about their potential regulative roles in important physiological processes.
Dedicated to research in non-coding RNAs (ncRNAs) for a decade, CHEN and YANG took note of this special type of ncRNAs quite early. In 2011, they developed a specific RNA-seq technology to profile nonpolyadenylated RNAs in a genome-wide scale and for the first time found stable nonpolyadenylated RNA-seq signals in specific intron and exon regions (Yang et al, Genome Biol, 2011). Later on, in September 2013, they systematically identified diverse circular RNAs originated from intronic sequences, and proposed for the first time their biogenetic mechanism as well as molecular functioning (Zhang et al, Mol Cell, 2013). The next year the teams unveiled a diverse family of circRNAs originated from extronic sequences and took the lead to propose a model called “alternative circularization” to explain in detail their ubiquitous genesis (Zhang et al, Cell, 2014). These results brought to light the fact that circRNAs universally exist in eukaryotic cells and signal complexity and diversity in gene expression regulation. Also, they raised the question what could be the implication of their ubiquitous expression in the cell.
Three years later, CHEN’s lab further discovered that circRNAs could bind to a series of protein factors, including NF90 and NF110, which are associated with immune responses to viral infections. Noting that more investigation into the functions of these circRNAs was required, CHEN’s team predicted that the binding of circRNAs to protein factors might involve important biofunctions in the cell (Li et al., Mol Cell, 2017).
Bridle on the Steed
Their prediction came true.
They discovered that circRNAs could suppress innate immune responses in the absence of viral infection by binding to a protein kinase, an immune receptor that plays a vital role in an antiviral defense pathway dominant in vertebrates. Once activated by certain double-strand RNAs (dsRNAs), this immune receptor can fight against the invading pathogen, and hence is termed dsRNA-activated protein kinase (PKR). Interestingly, PKR preferably binds to circRNAs over linear ones despite their identical sequences.
“Therefore,” the authors concluded, “this binding is dependent on some structural conformation rather than specific sequences.”
The researchers found in their experiments that most circRNAs tend to develop various short duplexes on their chains, just like a reel of earphone wires going wild. It is exactly these secondary structures that have given them the ability to bind and cap onto PKR, to suppress its phosphorylation and hence prevent its activation.
This latent warrior can be summoned and activated by certain types of long-chain dsRNAs. Paradoxically it stays quiescent when capped with circRNAs despite the latter’s double-strand structure. Their relatively shorter sequences might cause the difference, the researchers found. “Previous studies showed that activating PKR requires a dsRNA longer than 33 base pairs,” stated CHEN, “while shorter double-strand structures of 16 to 33 base pairs can block its activation instead.”
The duplexes developed on the rings of circRNAs contain only 16 to 26 base pairs, the authors noted. These relatively shorter lengths give them the specific ability to bind to PKR.
This temporary combination can break down at the early stage of a real viral infection. In this case, a nucleic acid receptor called RNase L responds to the viral stimulus and catalyzes the degradation of the inhibitor circRNAs, hence releasing the latent warrior PKR to fight against the invader.
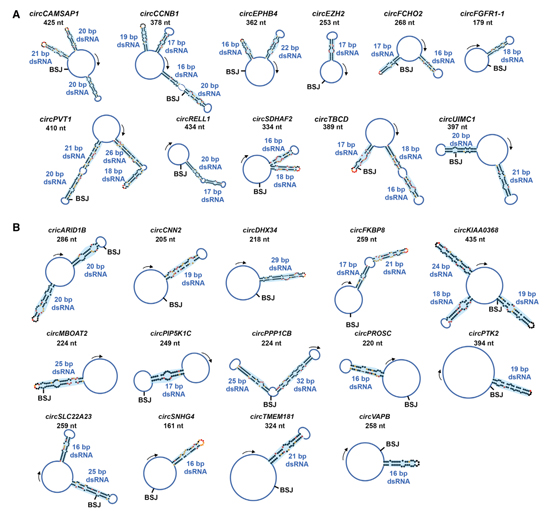
Various secondary structure models of 25 circRNAs as revealed by the researcher via different methods. The length of such circRNAs ranges from 16 to 26 base pairs:
(A) Secondary structure models of 11 circRNAs from in-cell SHAPE-MaP. Blue shadows show intra-dsRNA regions. BSJ sites and lengths (nt) of circRNAs are labeled. ?(B) Secondary structure models of additional 14 circRNAs with intra-dsRNA regions from in-cell SHAPE reactivities. (Image by CHEN’s lab, extracted from the Cell paper)

Dissolving “anchors:” Ring-shape RNAs, which lock on the natural immune factor PKR in normal circumstances, are illustrated “melting down” and hence releasing the latter for a fight against the invading viruses when a viral infection occurs.
The viral infection stimulates and activates a nucleic acid receptor called RNase L, also a protein kinase. It catalyzes the degradation of circRNAs and frees the warrior PKR from detainment. (Image by CHEN’s lab)
The low occurrence of back splicing plays an intriguing role here. Mistake is mistake – Mistakes happen at a rate less than 1% of the frequency of canonical splicing, though they occur globally in the cell (Zhang et al, Cell Rep, 2016). This leads to the relative scarcity of circRNAs compared to linear RNAs with the same sequences, though both of them can be degraded by RNase L. This makes it hard to restock this “mistaken product.” The insufficiency in inhibitors allows a high level of activity of the PKR, and hence it enters the antiviral battle with enough fighting capacity.
Astray Steed
Dangers like viral infections entail swift responses by the immune system. An immature immune response invoked at a wrong time, however, could cause disaster. An unfortunate example is SLE, in which the overly active immune system attacks the healthy cells of the patient, causing severe damage to the body.
Taking note of this, CHEN wondered if the overly active immune system is the result of a shortage of circRNAs. The ring-shape RNAs might help stabilize the immune system in normal conditions, she speculated.
To confirm her suspicion, she went to Dr. SHEN Nan, a leading expert of lupus in China, to discuss the matter. CHEN’s proposal of possible cooperative research was agreed to by SHEN. Hence an alliance was formed between the labs of CHEN and YANG and SHEN’s group at SIRRH, an affiliate of the School of Medicine of Shanghai Jiao Tong University. The joint team tested their speculation in a series of rigorous experiments and found that SLE patients do have a slight “overdose” of active RNase L in their immune cells. Further, they verified that this overdose of RNase L is highly correlated to an underdose of circRNAs. After artificially introducing circRNAs into immune cells originated from SLE patients, the researchers observed that the irritated PKR could be returned to a quiescent state, hence curbing the overreaction of the immune system.
This demonstrated that the slight overdose of catalyst RNase L has prompted a response in the patient and degraded a small number of circRNAs. This partially frees the PKR at a time when no viral infection is at work, triggering an unprovoked battle against a wrong enemy, an otherwise healthy body. A restocking of circRNAs helps curb PKR’s possible overreaction and reserves its power until the time comes for a fight against the right enemy.
“It’s an exciting discovery, as it has answered a very important question in research of this autoimmune disease pertaining to its most critical pathogenic pathway,” commented SHEN, director of SIRRH. Clinical immunologists have identified overly activated antiviral responses as the most important pathogenetic feature of such diseases, but they failed to determine the underlying mechanism. “For over 10 years we had been questing in vain for the reason why the responses spontaneously get activated, even when viral attacks are absent,” he said. “Now we have very exciting clues!”
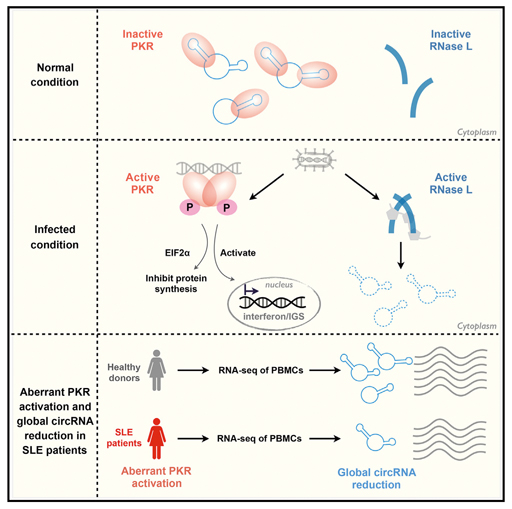
The joint team revealed in great detail the complete pathway mediated by circRNAs regulating the proper behavior of PKR, an immune receptor that meanwhile fights invading viruses. (Image by CHEN’s lab, extracted from the Cell paper)
Another highlight of this research is, SHEN commented, it could offer a novel specific and quantitative molecular marker for monitoring SLE disease activity and also a specific assessing criterion for its clinical treatment. “This has filled a long-existing gap,” he concluded.
The exact mechanism driving the spontaneous activation of RNase L in patients is still to be determined by further investigation, the authors noted. However, their findings have uncovered an unexpected pathophysiological connection of circRNA function to autoimmune diseases and hence offered new clues for future research into such diseases, experts say.
Reference
Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui YG, Dong KG, Ding HH, Qu B, Shen N*, Yang L*, Chen LL*. (2019) Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell, 177: 865–880.
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin QF, Wei J, Yao RW, Yang L*, and Chen LL*. (2017) Coordinated circRNA Biogenesis and Function with NF90/NF110 in Viral Infection. Mol. Cell, 67: 214–227.
Zhang XO, Wang HB, Zhang Y, Lu XH, Chen LL*, Yang L* (2014) Complementary Sequence-Mediated Exon Circularization, Cell, 159: 134–147.
Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL*. (2011) Genomewide characterization of non- polyadenylated RNAs, Genome Biol, 12: R16.
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu SS, Yang L*, Chen LL*. (2013) Circular Intronic Long Noncoding RNAs, Mol Cell, 51: 792–806.
Zhang Y, Xue W, Li X, Zhang J, Zhang JL, Yang L, Chen LL*. (2016) The biogenesis of nascent circular RNAs. Cell Rep, 15:611–624.


