By YAN Fusheng (Staff Reporter)
How a fertilized egg grows into a full-term mammalian embryo has fascinated scientists. It is incredible that one cell divides and differentiates into ~200 different types of cells, which later develop into vital organs and form a being. This well programmed differentiation has long intrigued biologists with a fundamental question: How and when does nature seal an embryonic cell’s fate – say, to grow into a fetus, or to become its placenta upon their first choice of fate? Now this question is partly answered by researchers at the Institute of Zoology (IOZ), Chinese Academy of Sciences (CAS).
Jointly led by CAS Member Prof. ZHOU Qi and Prof. LI Wei, the team reports that an unequally distributed long noncoding RNA, LincGET, can bias the first cell fate choice in mouse two-cell blastomeres, cells formed by the cleavage of a fertilized egg. One of the two-cell blastomeres that express higher level of LincGET tends to grow into the inner cell mass (ICM), which later becomes the fetus. This work updates our recognition on when this first cell fate becomes biased: the fate choice becomes biased earlier at the two-cell stage, rather than at the four-cell stage as previously reported. This work was published December 13, 2018 in the high-impacting journal of Cell.
Embryonic cells face their first fate choice at the two-cell blastomeres, cells formed by cleavage of a fertilized egg, owing to an unequally distributed long noncoding RNA, LincGET. The one and its progeny cells with higher numbers of LincGET tend to grow into the fetus and that with lower numbers of LincGET tend to form placenta. ? Prof. LI Wei & ZHOU Qi, IOZ, CAS.
To Be, or Not to Be: That Is the Question
To form an early embryo, a mammalian fertilized egg undergoes multiple cleavages through two-, four-, eight-, sixteen-cell blastomeres by order. The blastomeres face their first fate choice by either turning into the inner cell mass (ICM), which contributes to the fetus, or the trophectoderm (TE), which contributes to the placenta.
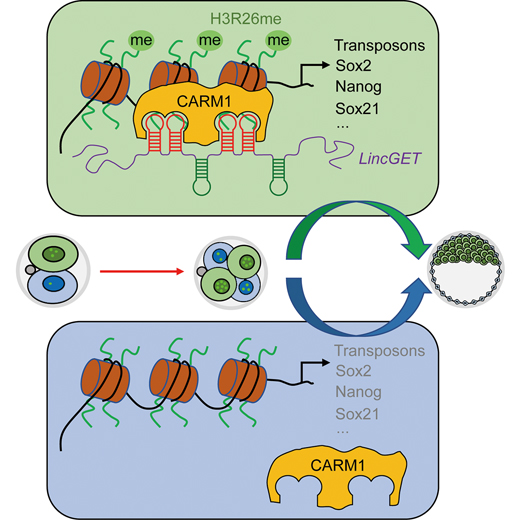
How do the cells make their first fate choice? In essential, molecular heterogeneity – molecular differences in a cell that cause different genes to be expressed – is the driving force for this bifurcation, to become the fetus or to form the placenta. Using mouse embryos as the model, former researchers found that molecular heterogeneity appears as early as at the four-cell stage: a protein called CARM1 distributes unequally within the four-cell blastomeres and biases the cell fate towards ICM. This finding intrigues ZHOU and LI: Could it be possible that the observed heterogeneity at the four-cell stage actually has arisen sometime earlier? Maybe some factors from the two-cell stage have already decided this upcoming heterogeneity.
Drawing on data from the single-cell RNA sequencing of many two-cell blastomeres, ZHOU and LI discovered a new molecular heterogeneity within two-cell blastomeres. Specifically, a long noncoding RNA, LincGET, expresses unequally within the late two-cell to early four-cell blastomeres, suggesting its potential role in biasing cell fate. Based on this assumption, they found that overexpression of LincGET in one of the two-cell blastomeres biased its progeny cells toward the ICM fate; while knocking down of LincGET blocks this tendency. Therefore, they inferred LincGET as the earliest molecule, so far documented, that biases cell fate in two-cell blastomeres.
How LincGET Exerts its Power
To seal the embryonic cell’s fate, the researchers found that LincGET recruits CARM1 – a protein known to bias the cell fate into ICM – to increase chromatin openness and activate ICM-specific genes. This conclusion was drawn on many converging evidences.
They first found that LincGET physically overlaps with CARM1 in cell nucleus, suggesting their potential interactions. To confirm this possibility, they designed two elegant “molecular fishing” tests to check whether CARM1 comes along when tagged LincGET is specifically fishing out from the two-cell-stage embryonic lysate. The data showed that CARM1 sticks to the bait together with LincGET in both tests, affirming their interactions.
By overexpressing LincGET while knocking-down CARM1 in one of the two-cell blastomeres, they found that once CARM1 is in shortage, a high level of LincGET alone will fail to exert its biasing power. Similarly, truncated versions of LincGET omitting its CARM1-binding sites also lose the biasing power while CARM1 presents itself normally. Hence, LincGET exerts its biasing power by recruiting CARM1 into function.
They also sought to understand how the interacting LincGET-CARM1 complex causes the cell fate bias. Drawing on the data that rates chromatin openness, they found that this complex enhances chromatin openness for the ICM-specific genes, while it induces the TE-specific genes less open. Notably, high chromatin openness of a gene (or more specifically its promoter, which is like ‘The Registration’ that allows the transcription machinery to check-in and spurs them into work) correlates with high gene expression.
Though how this RNA-protein complex enhances chromatin openness remains unclear, the researchers suggested that it could be mediated by activating transposons, jumping DNA segments capable of changing positions in genome. Interestingly, ICM-specific genes locate near transposons. The jumping transposons may disseminate their active chromatin status towards the neighboring ICM-specific genes. Nevertheless, how the RNA-protein complex activates transposons remains to be explored. Intriguingly, the jump of transposons may cause DNA mutations, which could be passed onto the germ line (sperm or egg cells) and thus also the species gene pool. In other words, LincGET may also have played a role in mammalian evolutions.
Implications
Summing up, the team demonstrated that as early as at the two-cell blastomeres, a cell has made its own fate choice, and unraveled the major scenario of its underlying mechanism. A nuclear long noncoding RNA, LincGET, was found to play a key role in making this initial fate choice: it recruits a down-stream protein to form an RNA-protein complex that increases chromatin openness, activates ICM-specific genes and hence predisposes the cell fate towards ICM.
“This finding helps us understand the transition between totipotency and pluripotency and how a cell’s fate is sealed in early embryos. It will also provide guidelines for producing stem cells with higher pluripotency or even totipotency. Last but not least, it shall shed some light on the functional studies of other long non-coding RNAs in early embryos,” stated the researchers.
Indeed, latest evidence has shown that when two-cell blastomeres are separated, mostly, only one of them develops into a mouse, contradicting with the previous understanding that both the two-cell-stage embryonic cells are generally considered to be totipotent – the potency of developing into all types of cells, tissues, or extra-embryonic tissues like placenta. Yet how does this happen still puzzled scientists until recently. The findings from the current work concerning the uneven distribution of LincGET within the late two-cell blastomeres and its function in biasing lineage fate provide a molecular explanation for this long-standing puzzle.
After this first fate decision, the two-cell blastomeres grow into either fetus or placenta by losing their totipotency, and march towards pluripotency, holding the potency of developing into different types of body cells.

