By YAN Fusheng (Staff Reporter)
DNA is the language of life with the alphabet of four nucleobases – adenine (A), thymine (T), guanine (G), and cytosine (C). Language has the power to define and converse, and so does DNA. DNA stores genetic information and defines what a living thing looks like. DNA can also be interpreted into the molecules of RNAs and proteins that are the two key players to support a life and interact or “converse” with other living things.
In the 1980s, scientists established a new form of DNA language to link the DNA strands of certain sequences to a certain nanoscale pattern or shape. By doing this, scientists can actually encode the blueprint of a target construct in the sequences of its DNA building materials. This marks the advent of DNA nanotechnology.
In the infancy of DNA nanotechnology, repetitive patterns were readily produced via a ‘seed-growing’ method, where a basic DNA motif, or a seed, grows on its own accord when basic motifs are added to ultimately form a desired pattern (figure 1). It is, however, quite hard to curb the growing pattern to an exact size with this method.
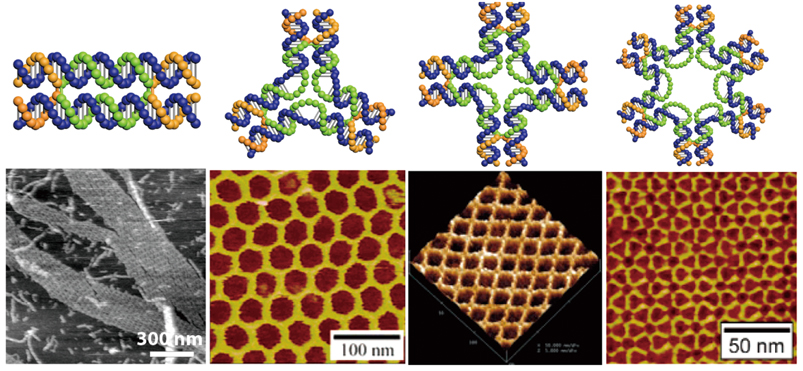
Figure 1 | Designs of some basic DNA motifs and patterns built from these basic motifs. The patterns are imaged with atomic force microscopy (AFM), a technique that scans the height of a probed area. (Credit: Hao Yan of Arizona State University)
It is also hard to encode DNA stands to construct a three-dimensional (3D) shape because this sort of shape-to-sequence translation is usually laborious. The hardship greatly scales up when it comes to constructing asymmetric and complex shapes.
As this field advanced forward, these limitations became obvious obstacles stopping DNA nanotechnology to fully bloom. Then, it came to the big question: How can we make an arbitrary shape out of DNA strands?
This question was very well answered in 2006 by Paul Rothemund, a computer and bioengineering scientist at Caltech, who came up with the idea to use numerous short single strands of DNA (the so-called “staples”) to direct the folding of a long single-stranded DNA (the “scaffold”) into a desired shape. He named, for the first time, such DNA folding as the “scaffolded DNA origami.” In a typical run, DNA strands of staples and scaffolds are mixed, then heated and cooled. As the DNA cools, the staples pull and fold the long scaffold DNA strand into a desired shape autonomously.
The autonomous nature of the folding process is governed by a strict rule called the Watson-Crick base pairing rule, where the DNA bases adenine (A) binds to thymine (T), guanine (G) binds to cytosine (C). A single-stranded genomic DNA (7,249 nucleotides in length) of M13 bacteriophage, a virus that infects and replicates within bacteria, is usually used as the scaffold where the sequence is already known. Therefore, it is the sequences of staples that actually define the final shape of a DNA origami. Because of this, Paul Rothemund wrote a computer program to enable an automatic output of staple sequences that are required to fold the scaffold into an exact shape, or virtually any shape.
To produce a desired shape in practice, images are drawn with a raster fill of a long single strand scaffold DNA. This framework is then fed to the computer program that calculates the placement and sequence of individual staple strands that bind to and fold the scaffold DNA into a desired shape (figure 2a).
Up to now, a variety of 2D or 3D shapes have been created with DNA origami (figure 2b-c). These designs can be directly observed with atomic force microscopy (AFM) – a microscopy that scans the height of a probed area and turns it into an image – or electron microscopy (EM), a microscope that uses a beam of accelerated electrons to illuminate minute objects.
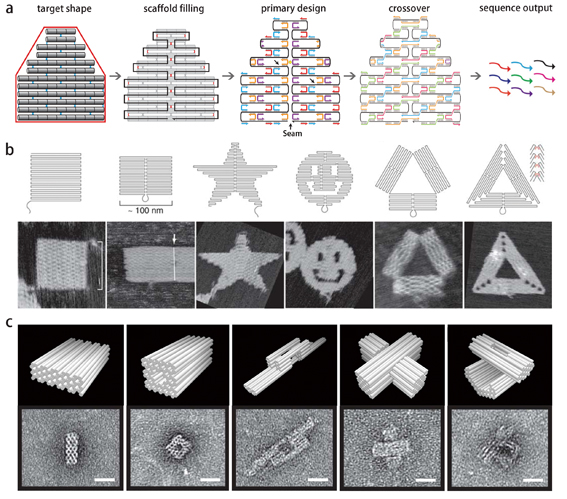
Figure 2 | Folding DNA into shapes. a) To produce a desired shape (outlined in red), images are drawn with a raster fill of a single long scaffold DNA strand. This design is then fed into a computer program that calculates the placement of individual staple strands. To hold the construct more tightly, the positions of nick (indicated by black arrows) are sealed with crossovers that transverse between the parallel helices. The middle seam is also bridged by merging staples in the primary design. Ultimately, the staple sequences in the final design are known following Watson-Crick base pairing, and outputted automatically. b) Designs of 2D shapes in raster fill and observations by atomic force microscopy (AFM), a technique that can feel and depict the height of a surface. (Credit: Paul Rothemund). c) Designs of 3D shapes in cylinder models and observations by transmission electron microscopy (TEM), a microscope that uses a beam of accelerated electrons to illuminate minute objects (Credit: William Shih of Harvard University).
As scientists demonstrated the possibility to construct various shapes with DNA origami, the next question would be how to put these shapes into use.
“The power to manipulate DNA in this way could change the way we make things at a very basic level,” stated Paul Rothemund at Caltech.
Indeed, the most powerful feature of DNA origami is the addressability that means a designer can precisely put objects of choice to exact positions. Because of this, scientists can unprecedentedly control the spatial locations of certain particles or molecules with nanoscale precision, which leads to numerous creative uses with DNA origami.
Here are some typical examples of how scientists make use of DNA origami. I hope, this could put you into perspective the amazing power of DNA origami at controlling and building things at a new level, as well as the creativity of scientists who put these designs into function.
DNA Nanotemplate-aided Fabrications
The addressability of DNA origami enables a precise control over the spatial distribution of particles or molecules when they are anchored to a DNA origami. Because of this, DNA origami can be used as a precise template for nanoscale fabrication.
For example, scientists used DNA rings of different sizes as the nanotemplates to generate and tailor liposomes – artificial membrane vesicles used especially to deliver drugs to cancer cells – to exact sizes (figure 3a). The production of liposomes with a uniform size has proven very hard to achieve via traditional methods. The production of liposomes at exact sizes offers the chance to find out the optimal size that allows the most efficient drug delivery to cancer cells in vivo.
Just like any expanding material can be shaped inside a mold by replicating the cavity, a growing inorganic nanoparticle can also be casted and shaped inside a nanomold formed by a DNA origami (figure 3b).
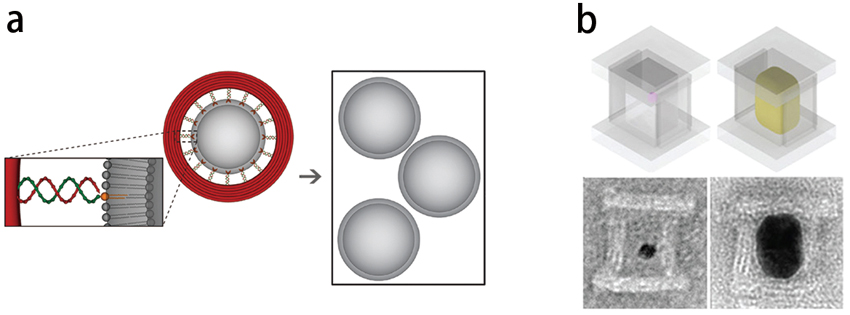
Figure 3 | DNA origami-based nanotemplating and nanocasting. a) A DNA ring is used to tailor liposomes into an exact size (Credit: Chenxiang Lin from Yale University). b) A box-like DNA nanomold is used to cast and grow nanoparticles to a defined shape (Credit: Peng Yin, Harvard's Wyss Institute).
This concept of ‘nanocasting’ mimics the way of growing cubic watermelons, by nurturing an individual watermelon seed to maturity inside a cubic glass box. Similarly, when being placed within an enclosed stiff DNA nanomold with a designer 3D cavity, a nucleating gold ‘seed’ can grow to full size that fills and replicates the cavity. Besides, this method can be generalized to cast other inorganic nanoparticles to arbitrarily shapes defined by DNA origami.
“Such a method may lead to computationally designed functional materials for the digital manufacture of optical nanocircuits, electronic nanocomputers, and perhaps even sophisticated inorganic nanorobots, each with their blueprints (or “genomes’’) encoded in the DNA molecules that constitute their nanofabricators,” stated Prof. Peng Yin from Harvard's Wyss Institute, who develops the nanocasting technique.
Smart DNA Nanorobots
DNA can also self-assemble into smart DNA nanorobots that can open in response to binding molecular keys. For example, Danish scientists created a DNA box with a smart lid that can be opened when a specific DNA strand is introduced as the key (figure 4a). This design exploits a duplex-gated lock mechanism that can be unlocked when one strand of the duplex is displaced by a strand of DNA key.
This duplex-gated lock mechanism can also be extended to sense the presence of protein keys. It can be achieved by replacing one strand of the duplex lock with a structure-switching aptamer, a type of protein-binding DNA molecules. Aptamer strands, when carefully designed and placed, can seal up a hollow origami structure and form a DNA aptamer-based lock mechanism that opens in response to binding protein keys. These protein keys can be highly expressed tumor markers either on tumor cells or within tumor surroundings. So, when the nanorobots enter into the tumor surroundings, they will be unlocked upon the binding of target proteins (the keys) to the aptamers (the locks). Once the nanorobots unfold themselves, the disclosed drugs can then set to work. Aptamer-gated DNA nanorobots can be used for targeted transport of therapeutic antibodies to tumor cells, as demonstrated by an in vitro study (figure 4b).
More recently, CAS researchers from the National Center for Nanoscience and Technology (NCNST) successfully devised smart DNA nanorobots that can precisely deliver thrombins – enzymes in blood that cause blood clotting – to the tumor site to cause blood blockage and starve tumor cells to death (read more on page 18-19) (figure 4c).
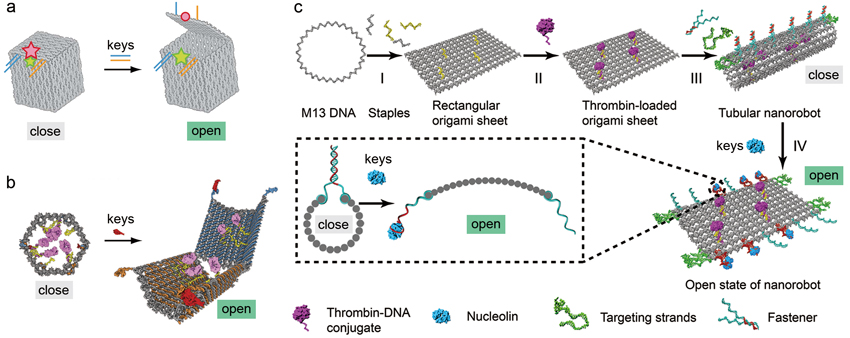
Figure 4 | Designs of smart DNA nanorobots. a) A smart DNA box that can be opened by DNA keys (Credit: Kurt Gothelf and J鴕gen Kjems, Aarhus University). b) An aptamer-gated DNA nanorobot unfolds itself upon the binding of protein keys and reveals its payload (Credit: George Church from Harvard Medical School). c) Design of an aptamer-gated smart DNA nanorobot for tumor infarction therapy. The DNA nanorobot can open upon the binding of nucleolin (the key) ?a surface protein that highly expressed on tumor-associated endothelial cells ?to the aptamer-containing duplex (the lock). Once disclosed, the thrombins can set to work by forming blood clots that cut the blood supply to tumor cells and stave them to death (Credit: DING Baoquan, CAS National Center for Nanoscience and Technology).
DNA Nanogadgets to Probe Biological System
Minute DNA origami-based nanogadgets enable scientists to probe biological system at an unprecedented resolution. Details that appear to be elusive previously, can be now approached. In other words, DNA nanogadgets enable scientists to perceive “the devil in the detail” and thereafter gain new insights.
For example, proteins, when precisely patterned on a DNA origami, can mimic the patterns of membrane proteins on cell surface. These DNA nanogadgets allow scientists to test a long-standing hypothesis – cells could read patterns of membrane proteins on neighboring cells and then react differently and accordingly to these different patterns.
“You could think of it as the cells having a kind of ‘tactile alphabet’, like the Braille system for blind reading. But instead of recognizing dots on a paper, cells read patterns of proteins on neighboring cells. It has been a challenge to actually test this hypothesis and examine this tactile alphabet of cells because it was always very hard to pattern proteins at nanoscale precision and even harder to do it in suspension (i.e. not patterned on a surface),” stated Bj?rn H?gberg, a Swedish scientist who has a good skill at making DNA nanogadgets, “With DNA origami we can make very precise patterns of proteins ligands and study the effect the patterns have on receptor activation.”
To test this hypothesis, Bj?rn H?gberg and his coworkers chose to study two interacting membrane-bound proteins, ephrin ligands (ephrins) and Eph receptors that bind to each other to relay a direct cell-cell communication. They devised ‘ligand nanocalipers’ where the ephrin ligands are spatially distributed at a rod-like DNA origami with nanoscale precision (figure 5a). Using these nanocalipers, they showed that the nanoscale spacing of ephrin-A5, one type of ephrin ligands, directs the activation of EphA2, a type of Eph receptors, in human breast cancer cells. In particular, ephrin-A5 nanocalipers of 40-nm spacing (NC40) can more efficiently activate EphA2 compared to that of 100-nm spacing (NC100). Besides, these spatially distributed ephrin ligands were shown to tune the invasive properties of the breast cancer cells.
Ligand nanocalipers may also offer a chance to fathom another long-standing puzzle of why targeting a plasma receptor could simultaneously lead to tumor-promoting and tumor-suppressing effects. The knowledge gained from such studies may provide new insights for a rational drug design.
More recently, Bj?rn H?gberg and his coworkers designed another DNA nanogadget to probe the immune system – how antibodies bind and dynamically interact with spatially patterned antigens (figure 5b). They precisely patterned antigens (foreign substances that trigger immune responses of the host) at various separations on a DNA origami to study how the different separations affect the binding of antibodies, Y-shaped antigen-binding proteins where each arm can hold to an antigen. By doing this, scientists sought to understand how the repetitive patterns of antigens relate to the initiation of immune responses.

Figure 5 | DNA nanogadgets. a) Ligand ‘nanocalipers’ with precise spacing of ephrin ligands on a DNA nanorod to probe the effect the patterns have on receptor activation. b) Nanopatterned antigens on a DNA origami to probe the spatial tolerance of binding antibodies. (Credit: Bj?rn H?gberg, Karolinska Institutet)
The spatial arrangements of antigens were thought to be relevant to certain immune responses. In fact, this concept has been successfully exploited for vaccine development through displaying antigens in patterns. Exactly how a certain pattern elicits a strong response, however, is poorly understood. A deeper knowledge concerning the dynamics of antibody binding to variably distributed antigens would present itself as an important missing hinge in the chain of immune reaction. In particular, no methods have been able to show precisely the optimal antigen separation required for the most stable binding by an antibody.
Using these precisely patterned DNA nanogadgets, the team found that all studied antibodies can bind bivalently to two antigens separated at distances that range from 3 to 17 nm. Notably, a separation of approximately 16 nm yields the most stable bivalent binding. This newly acquired knowledge may enable scientists to design better vaccines or antibodies to tackle with various disease conditions, such as virus infection and cancer.
Future Prospects
DNA has become one of the most widely used building blocks for producing desired shapes with nanoscale precision. The field of DNA nanotechnology has attracted an influx of non-biologists, including physicist, computer and engineering scientists, over the last decades. Apart from what has been enlisted in this piece, DNA origami has also demonstrated great potential in DNA computing, super-resolution microscopy and designer optimal devices. With the continuous influx of brilliant minds and the intercrossing of disciplines, a full bloom of DNA nanotechnology is on the horizon.
References
1. Paul W. K. Rothemund, Folding DNA to Create Nanoscale Shapes and Patterns. Nature 440, 297 (Published: March 01, 2006). doi: 10.1038/nature04586.
2. Y. Yang, J. Wang, H. Shigematsu, W. Xu, W. M. Shih, J. E. Rothman, C. Lin, Self-Assembly of Size-Controlled Liposomes on DNA Nanotemplates. Nature chemistry 8, 476 (Published: March 21, 2016). doi: 10.1038/nchem.2472.
3. W. Sun, E. Boulais, Y. Hakobyan, W. L. Wang, A. Guan, M. Bathe, P. Yin, Casting Inorganic Structures with DNA Molds. Science 346, 1258361 (Published: Nov 7, 2014). doi: 10.1126/science.1258361
4. E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, . . . J. Kjems, Self-Assembly of a Nanoscale DNA Box with a Controllable Lid. Nature 459, 73 (Published: May 7, 2009). doi: 10.1038/nature07971.
5. S. M. Douglas, I. Bachelet, G. M. Church, A Logic-Gated Nanorobot for Targeted Transport of Molecular Payloads. Science 335, 831 (Published: February 17, 2012). doi: 10.1126/science.1214081.
6. Suping Li, Qiao Jiang, Shaoli Liu, Yinlong Zhang, Yanhua Tian, Chen Song, . . . Yuliang Zhao, A DNA Nanorobot Functions as a Cancer Therapeutic in Response to a Molecular Trigger in Vivo. Nature biotechnology 36, 258 (Published: February 12, 2018). doi: 10.1038/nbt.4071.
7. A. Shaw, V. Lundin, E. Petrova, F. Fordos, E. Benson, A. Al-Amin, A. Herland, A. Blokzijl, B. Hogberg, A. I. Teixeira, Spatial Control of Membrane Receptor Function Using Ligand Nanocalipers. Nature methods 11, 841 (Published: July 06, 2014). doi: 10.1038/nmeth.3025.
8. A. Shaw, I. T. Hoffecker, I. Smyrlaki, J. Rosa, A. Grevys, D. Bratlie, I. Sandlie, T. E. Michaelsen, J. T. Andersen, B. Hogberg, Binding to Nanopatterned Antigens Is Dominated by the Spatial Tolerance of Antibodies. Nature nanotechnology 14, 184 (Published: January 14, 2019). doi: 10.1038/s41565-018-0336-3.

